In search of endogenous proteins
Johannes Broichhagen
www.broichhagenlab.com
In his lectures, summarized in his book “What is Life?”, the physicist Erwin Schrödinger described in the 1940s that “the course of life processes in an organism […] [shows] an admirable regularity and order that in inanimate matter does not find its equal”.[1]
Almost 80 years later, we are still searching for the origin and description of “life”, and with the help of molecular biological (e.g. gene editing using CRISPR/Cas9), mass spectrometric (e.g. proteomics, metabolomics), high-throughput Using sequencing (e.g. single cell RNA sequencing) and high-resolution microscopic (e.g. super-resolution STED and STORM) techniques, we try to put together the many pieces of the puzzle that have been found into a regular and orderly overall picture.
Scientists are not only driven by the most fundamental question about life, but in particular by the connections and causes in health and disease patterns in order to find better and safer therapies and medicines. A large body of knowledge has been created through the use of numerous biological model systems, but key questions remain difficult to answer, such as:
- “How many proteins of one type are there per cell?” and
- “How are they spatially distributed at a given point in time and depending on cell activity?”
These are interesting questions, especially when dealing with non-model systems, such as biopsies from patients, that are not (or very difficult) to genetically manipulate. In order to get a deeper insight into the world of biomolecules, their interactions, localizations and complexity, our research group deals with the design and use of tailor-made probes at the interface of synthetic chemistry and applied biology in the research field of Chemical Biology.
Richard Dawkins summarized a scientific task in 2013 with the words “Complexity is a problem that any theory of biology has to solve”[2]. At the cellular level, this is a complex orchestra of mainly proteins, sugars, fats and nucleic acids that control each other through their interactions. Peter Kruse names different (better and worse) solutions for how to react to growing complexity and describes impressively: “You can simplify a complicated system by trivializing it by subdividing it. You destroy a complex system if you trivialize it.”[3] So what exactly does this mean for our research?
We want to work as closely as possible to a complex biological system like the cell, and have formulated a few important criteria for this: (i) the investigation and manipulation of biomolecules in living (ideally primary) cells (ii) without the use of genetic engineering methods, which can lead to perturbations (iii) using non-invasive methods. The selection of the method determines the approach with a minimum of trivialization.
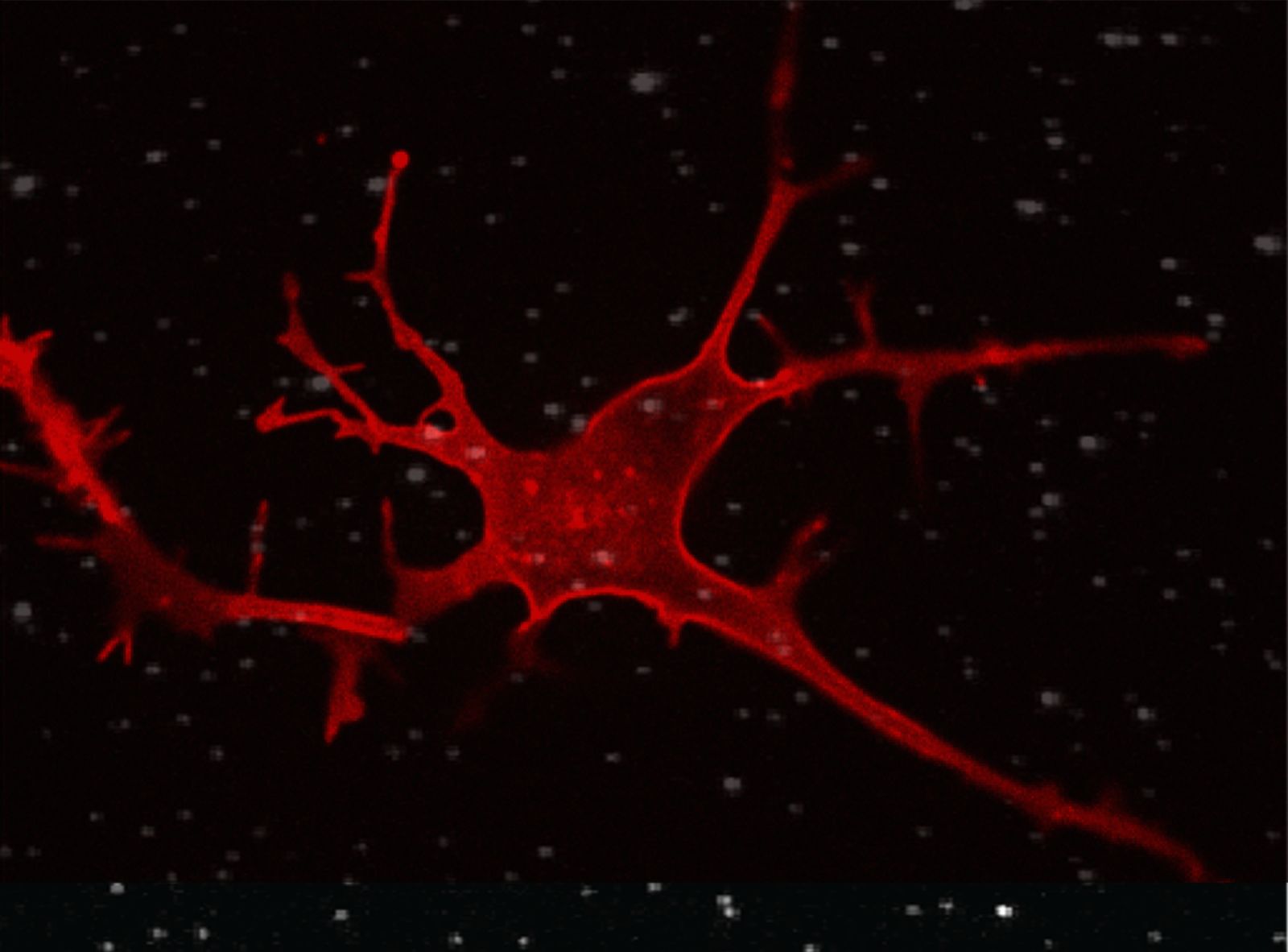
In contrast to other imaging methods such as ultrasound or magnetic resonance imaging, fluorescence microscopy ushered in the super-resolution age by being able to circumvent Abbe’s law and has since experienced a revolution. Using stimulated-emission depletion (STED) or stochastic reconstruction (STORM) microscopy, cellular resolutions of up to 20 nanometers can be achieved (for comparison: a cell nucleus has a diameter of 5-15 micrometers, a globular protein ~5 nanometers). Both systems require the introduction of fluorescent dyes, and it is hardly a coincidence that the development of these was recognized with the 2014 Nobel Prize in Chemistry.
To label proteins specifically, fluorophores must be attached to a molecular scaffold that recognizes its target precisely. Traditionally, antibody-fluorophore conjugates are used for this. However, even with a certain size, these are not so suitable, and function mainly in fixed cells or tissues, i.e. no longer in living systems. We try to produce individually designed probes with modern synthetic chemistry, based on validated pharmacology of active substances and with the help of crystal structures. These probes find their target molecule specifically and can be visualized by microscopy through the fluorophore linked to them.
This all works without genetic manipulation, and thus remains as close as possible to the cell’s native system. Furthermore, the development of dyes is a central research topic, which goes hand in hand with the development of probes. To cite one example: the introduction of deuterium, a hydrogen isotope, into rhodamine-based dyes shows an improvement in photophysical and chemical properties; the chromophores are brighter and less prone to bleaching.[4,5] This also works wonderfully in high-resolution STED microscopy, which places some demands on fluorophores due to the high laser intensities used.
As a second example, the LUXendins should be briefly presented.[6,7] These are specific, antagonistic peptides that bind to the glucagon-like peptide-1 receptor (GLP1R) without triggering a physiological response (Figure 1A). This makes them ideal as probes. Using bioorthogonal linker chemistry, these peptides could be decorated with dyes ranging from green to near-infrared. This enables the identification of this important receptor, which is pharmacologically targeted in the treatment of diabetes, and allows for a variety of exciting experiments. In addition to super-resolution microscopy, which resolved the nanodomains of this receptor, GLP1R could be found in various tissues, such as the pancreas and brain, and exciting single-molecule imaging could be performed, in which a receptor can be tracked as it moves on the cell surface (Figure 1B).
With the further development of such samples, we want to enlighten the fine orchestra of cell communication in the future, and for this we have laid a basis at the interface of organic chemistry and cell biology. To complete the circle with Erwin Schrödinger, the metaphor of his cat (Schrödinger’s cat) is very well suited: you only know whether the cat is alive or dead when you open the lid of the box in which it is sitting. So we only know something about some states when you can look them up – and in order to be able to look them up, we are developing new and smart tools.
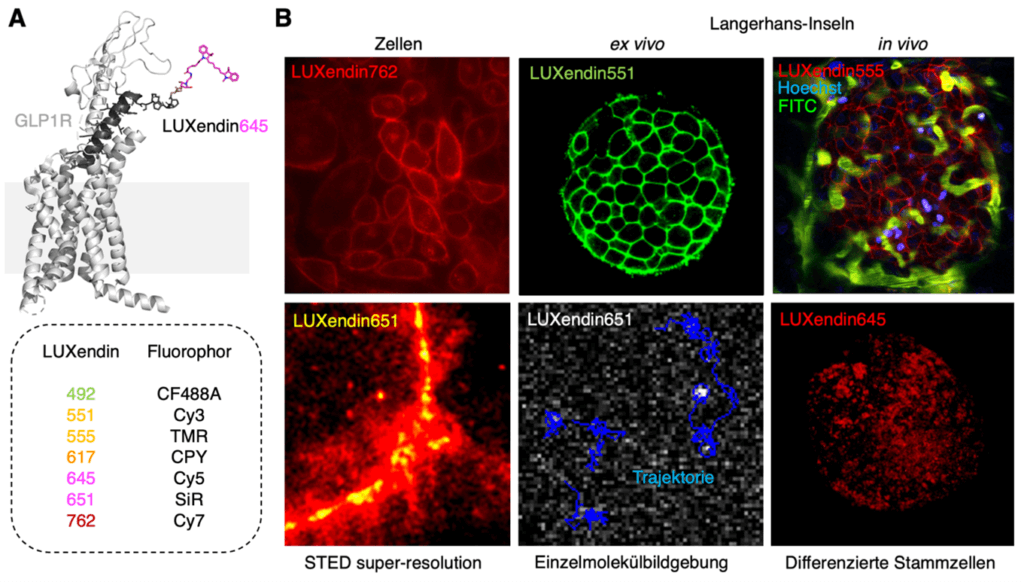
Figure 1: GLP1R labeling using LUXendin. A) LUXendin645 consists of a high-affinity fluorophore-linked peptide that specifically binds to the glucagon-like peptide-1 receptor (GLP1R) and can be colored. B) Various fluorescence microscopy images of GLP1R using LUXendins, in cells, islets of Langerhans (a mini-organ in the pancreas), for high-resolution STED microscopy and for single-molecule imaging.
References
[1] E. Schrödinger, „Was ist Leben?“ Piper Taschenbuch, 1989.
[2] R. Dawkins, 2002, https://www.ted.com/talks/richard_dawkins_militant_atheism
[3] P. Kruse, 2008, Interview im Rahmen der SCOPE_08.
[4] Grimm et al., JACS Au 2021.
[5] Roßmann et al., bioRxiv 2020.
[6] Ast et al., Nat. Commun. 2020
[7] Ast et al., JACS Au 2022.





 4c media
4c media